SupaVillain
Hazard to Others
  
Posts: 171
Registered: 28-10-2014
Member Is Offline
Mood: No Mood
|
|
Peptide Synthesis Help
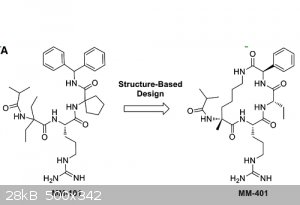
I need the name and procedure of an (likely aliphatic?) 4-carbon linker used to cyclize compound MM-101 into MM-401. They provide all info except for
this one part. The compound is made using 3mer peptide "Ac-ARA-NH2" and the bottom of page 47 of the document in the second link below show more
steps. The first link shows a pic of MM-101, next to a pic of MM-401, also attached.
pic of molecules here: http://www.cell.com/molecular-cell/fulltext/S1097-2765(13)00869-1
detailed paper about the molecules (go to bottom of page 47 for MM-101, top of page 49 for MM-401): liztown_2.pdf found here https://deepblue.lib.umich.edu/handle/2027.42/96011
at bottom of page 54 they say "We then generated a cyclic derivative of MM-101, MM-401, that has a 4-carbon linker connecting the N- and C- termini of
the peptidomimetic."
Thank you for any help
Oh.
|
|
|
ziqquratu
Hazard to Others
  
Posts: 385
Registered: 15-11-2002
Member Is Offline
Mood: No Mood
|
|
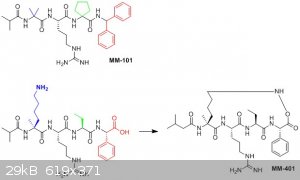
Forgive me if I'm misunderstanding your question, but it sounds like you're asking "how do they turn MM-101 into MM401" - that's not what they're
doing. From what I can see, they've previously studied MM-101, then based on its properties designed MM-401 and prepared it entirely separately. If
you look at them, you can see there are huge structural differences between the two.
For MM-401, they've got an a-methyl lysine in place of the a,a-diethyl glycine found in MM-101 - it's the lysine side chain that provides the
four-carbon linker (shown in blue, below). They've also replaced the cyclopentane system with an ethyl group (green), and the diphenylmethyl amide
with phenylglycine (red) - which is the other side of the cyclic system in MM-401.
I'd guess that they would synthesise the peptide linearly as shown at bottom left, then the ring would be formed by coupling the a-methyl lysine side
chain with the phenylglycine using a standard carbodiimide-based coupling reagent (which would probably be similar to the one used for the peptide
synthesis itself).
Oh, and please forgive the absurd C-N-O bonds in my scheme - I've done it that way to emphasise the point as to where the ring is coming from!
|
|
|
SupaVillain
Hazard to Others
  
Posts: 171
Registered: 28-10-2014
Member Is Offline
Mood: No Mood
|
|
So they still use Ac-ARA-NH2? Being Ac-alanine-arginine-alanine-NH2... Can you give me the steps at which this would be made? Or recommend the ethyl
group and a carbodiimide-based coupling reagent?
Oh.
|
|
|
ziqquratu
Hazard to Others
  
Posts: 385
Registered: 15-11-2002
Member Is Offline
Mood: No Mood
|
|
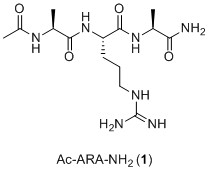
Sorry, Supa, I've only skimmed, but I'm not even sure where the "Ac-ARA-NH2" is coming from. That is NOT the sequence of MM-101 (arginine
is the only "natural" amino acid in that molecule - there is no alanine in either of them). From skimming, here's my bet - they found a large protein
or peptide and did mutation studies to figure out that the minimum binding motif was Ac-ARA-NH2 (compound 1, below). Then
they played around - trial and error, molecular modelling, or whatever - and found that MM-101 was better (stronger binding, say) than
Ac-ARA-NH2. Now they've kept going with their modifications and discovered MM-401, which is better (in some way) than either of the first
two compounds. If you compare compound 1 to either MM-101 or MM-401, you should see that while there are some basic similarities,
there are many very substantial differences between them.
Of course, this story I've told is pure speculation based mostly on other projects I've seen develop in this way and a cursory skim of this work, so
take it with a grain of salt! I'm sure a careful reading of the papers you linked would give a much better idea of the true background story.
So, for MM-401, here's how I'd go about it. Solid-phase peptide synthesis would give the linear precursor 2 (note - "PG" means
"protecting group", which stops the side chains from reacting when you don't want them to. Fmoc is also a protecting group, so that you don't simply
get polymerisation of the amino acid in solution, instead of coupling to the chain you're trying to make). The individual amino acids would either be
commercially available or prepared by one of any number of possible methods - a proper literature search would be required to sort that out.
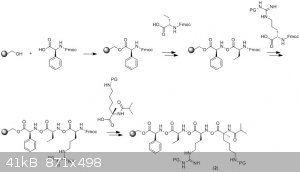
You would then cleave that peptide from the resin and remove the protecting group from the lysine side chain (you might also deprotect the arginine,
I'm not sure, but for the sake of argument I'm going to leave that as it is). Then you'd cyclise the lysine amine onto the C-terminal carboxylic acid,
followed by removal of the arginine protecting group, to give MM-401.
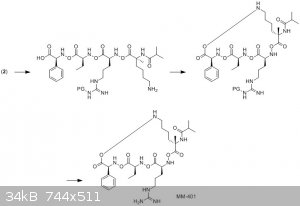
Hopefully that makes things somewhat clearer. As for the reagents you'd use; well, personally, I'd use Wang resin as my solid support, and then do the Fmoc-based peptide synthesis using a coupling reagent like HATU or EDC/HOBt (see also this paper for a discussion of some of the coupling reagents used in peptide synthesis). The deprotection steps would depend on the protecting
groups used, and then the cyclisation would use a similar coupling reagent to those I mentioned above - perhaps some trial-and-error to figure out
which ones give the best yield, if that's your goal.
---
hope this has cleared it up a little for you; let me know if something in particular is unclear (or, indeed, if I've completely confused you!). Also,
perhaps if you could give some insight as to your interest (are you just trying to understand something you came across, is this something you're
trying to replicate, etc.) I might be able to better tailor my answers.
|
|
|
SupaVillain
Hazard to Others
  
Posts: 171
Registered: 28-10-2014
Member Is Offline
Mood: No Mood
|
|
MM-401 is just outstandingly interesting, because it induces pluripotency in skin cells and some types, converting them into stem cells. Not only does
it do this, but it makes "naive" stem cells. Others have poor performance because, for example, a skin cell is prepared to be skin, not to be muscle
or nervous, so they won't perform well enough. However, naive pluripotency means they are returned to a state before that, and can truly be used to
become any other kind of cell.
I thought the "ARA" in Ac-ARA-NH2 stood for alanine-arginine-alanine, according to this chart.
Basically I got that from the liztown_2.pdf 32MB file from here.
I can easily read chemical syntheses but I am totally new to peptide synthesis. I at least found instructions on how to combine fmoc with the amino
acids. How can I learn the reasoning behind your selection of specific chems?
Oh.
|
|
|
Dr.Bob
International Hazard
    
Posts: 2656
Registered: 26-1-2011
Location: USA - NC
Member Is Offline
Mood: No Mood
|
|
The synthesis is not trivial, as using the best mix of protecting groups to allow the Lysine to be deprotected and then coupled to the Phenylglycine
will take some care, and Arg is a pain in most peptides. Since three of the amino acids are non-natural, the chemistry will not be easy or cheap,
this material could easily cost over $100/mg to make in small amounts. It does say that the synthesis of the MM-401 will be reported elsewhere, so
it will likely be printed in the next year or so, plus, they are likely patenting it, so that may be part of the delay. But there is no sign of it
being printed yet. If you really want to try to make this, find a book on peptide synthesis and learn about the protecting groups like Fmoc, Boc,
and Cbz, as they all might be used in this. It might even be a candidate for solution phase synthesis, since it is only 5 aa's long and cyclized.
I have made things like that before, but that would be about 10+ steps using pretty expensive reagents. I would protect the Lys with the isoBut
amide first, that would allow the gamma amine to be Boc protected, which will simplify the chemistry some.
|
|
|
SupaVillain
Hazard to Others
  
Posts: 171
Registered: 28-10-2014
Member Is Offline
Mood: No Mood
|
|
I have done an insane amount of digging, couldn't find the U-M patent or Ascentage Pharma license, might email the "leader of the team" who designed
MM-401, but I think I might have found its synthesis here at page 33 without its name even listed. The molecule looks identical to the one in liztown_2.pdf. I don't think I'll import all the lewis
structure images here, anyone who wants to see them can find them there at the link. I wonder if the writing at the bottom of my attached pic is the
synthesis in word form? This is like finding the secret krabby patty recipe.
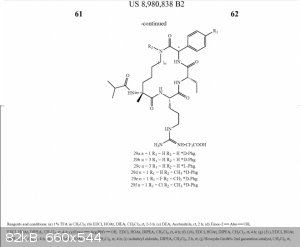
[Edited on 14-9-2016 by SupaVillain]
Oh.
|
|
|
ziqquratu
Hazard to Others
  
Posts: 385
Registered: 15-11-2002
Member Is Offline
Mood: No Mood
|
|
I believe you've found it, SupaVillain - that's certainly a viable procedure, and solves at least one of the problems associated with my proposal
(selective deprotection of the lysine residue I proposed is avoided because they don't use lysine - instead, they form the ring via olefin
metathesis). Of course, you still have many of the issues Dr.Bob noted, like the need for expensive unnatural amino acids, but it'd be easy enough
once you have them (and I know one - phenylglycine - and bet another - ethylglycine - are commercially available anyway).
Obviously it's a very brief overview of the method that they've provided; that said, it's enough to enable anyone reasonably familiar with the
chemistry to come up with a viable procedure. I'd be happy to help you understand what's going on, and even to help you design a method, if you're
having difficulties with it, but you suggested that you may be sufficiently familiar with organic synthesis to be able to understand the schemes on
the preceding pages. Just note that they appear to be reporting two different routes - one starting with a solid-phase synthesis, the other being
exclusively solution phase (perhaps because certain analogues weren't compatible with one or other of the methods) - which makes it a little more
difficult to follow.
|
|
|
Dr.Bob
International Hazard
    
Posts: 2656
Registered: 26-1-2011
Location: USA - NC
Member Is Offline
Mood: No Mood
|
|
That patent looks correct, they just bury the good stuff in the fluff. The pages 56-57 and a few past that are all relevant. If you know solution
phase chemistry, I would tend towards that path, as the resins based route takes a lot of special stuff. I do have some nice peptide chemistry
glassware if you decide to go that route, I can sell for it a very low price.
Good luck.
|
|
|
SupaVillain
Hazard to Others
  
Posts: 171
Registered: 28-10-2014
Member Is Offline
Mood: No Mood
|
|
There's special glassware just for solution phase synthesis? I really don't know anything about peptide synthesis other than the concept that
protection groups make reactions occur where you want them to, so you can make a chain, and then you remove them....
Oh.
|
|
|
Dr.Bob
International Hazard
    
Posts: 2656
Registered: 26-1-2011
Location: USA - NC
Member Is Offline
Mood: No Mood
|
|
You typically use a vessel with a joint or screw top on the top, and frit on the bottom, then a stopcock, to make it easy to wash the resin at each
step. And you normally rotate the vessel, rather than stir with a magnet, as that tends to grind the resin into dust. There are many variations,
but automated peptide synthesizers have a similar type of vessel, often a column with a frit on the bottom, and a valve below that, then chemicals are
squirted in at the top.
Some good examples are in the list below, but other companies make 100's of variations. If there is a joint at the bottom, it is easier to pull a
vacuum to rinse the resin quickly, or you can use a straight tube with a stopper. The Luer lock ones fit into manifold for easy rinsing.
http://www.chemglass.com/search_category.asp?category=C27&subcategory=(All)
I used to do that type of work many years back, haven't done much lately, but still use the vessels occasionally for work with other resins, there are
many resin bound reagents and scavengers which can help run reactions or purify chemicals easily. Most cost a lot, however.
|
|
|
SupaVillain
Hazard to Others
  
Posts: 171
Registered: 28-10-2014
Member Is Offline
Mood: No Mood
|
|
Do you actually know how much hoveyda grubb's 2nd gen catalyst I would need per an example amount of end peptide? It's like $70 for 100mg.
Oh.
|
|
|
SupaVillain
Hazard to Others
  
Posts: 171
Registered: 28-10-2014
Member Is Offline
Mood: No Mood
|
|
Turns out cheapest route for hoveyda grubbs is starting with a gram of alfa aesar's Dichlorotris(triphenylphosphine)ruthenium(II) and
Tricyclohexylphosphine, along with the other one listed on the grubbs catalyst wiki, and the (also written there) addition of an NHC and copper (I)
chloride to the 2nd gen Hoveyda grubbs. The patents cited in text of the wiki really illustrate it best.
HOAt, I am yet to review but molbase has its synthesis routes. All the others are far easier to get info on and make.
Still don't know how to find out how much to use though, don't know how stoichiometry works for peptides.
I hope ruthenium could be retrieved from used peptides and reactions because it is expensive.
Oh.
|
|
|
ziqquratu
Hazard to Others
  
Posts: 385
Registered: 15-11-2002
Member Is Offline
Mood: No Mood
|
|
I'd argue that you're looking for trouble trying to prepare those reagents...
Grubbs' II catalyst, as I recall, is one that requires truly careful anhydrous and oxygen-free techniques to prepare. The time and the cost of
reagents and equipment needed to make it might easily overrun the cost of the catalyst - $70 for 100mg isn't at all bad, as far as those types of
reagent go, and because your peptide has a high MW, you're only going to need a very small amount of catalyst (typically around 5 mol%, if memory
serves, for that type of metathesis) to get a lot of product.
Recovery of the waste ruthenium would be possible, I'm sure, but I'm not sure it would be economically viable on the scale you're talking about. If
you wash the post-metathesis solution with H2O2, you should be able to precipitate the ruthenium as RuO2 (I think);
check out this paper.
As for HOAt - again, I'd be loathe to synthesise such a reagent. If you can't get it, look at some alternatives - HOBt and N-hydroxysuccinimide (NHS)
are both common and cheap, and would likely work at least nearly as well as HOAt (you might take a small hit to your yields, but I wouldn't expect an
enormous difference for such a small peptide).
Finally, as to your unfamiliarity with peptide synthesis - you keep addressing it as if it were some weird discipline totally foreign to organic
chemistry; in reality, it's just a specific subset of organic chemistry! Peptide synthesis, at its core, is little more than making amides. All the
complexity comes about from trying to maximise yields; or deal with protecting group strategies; or handle compounds which, due to their side chains,
have properties which complicate purification. All of the functionalisation is just organic chemistry on slightly bigger molecules than most of us
deal regularly with. And, in this case, all of those problems have largely been addressed by the original authors. You might need to tweak the
procedure to accommodate the reagents you have available to you, but most of the various reagents used for peptide synthesis are largely
interchangeable without having a dramatic effect on the outcome. Heck, the choice of expensive HOAt over, say NHS may well be no different to the
choice you might make between, say, NaOH or KOH to neutralise an acidic solution - whichever one is closest to hand, whichever you happen to have more
of, or whichever one you've used most often in the past!
|
|
|
Dr.Bob
International Hazard
    
Posts: 2656
Registered: 26-1-2011
Location: USA - NC
Member Is Offline
Mood: No Mood
|
|
I agree, if going this route, buy the Grubbs cat, only need a small amount, but must be careful. You can almost always lower the amount of catalyst
and lengthen the time fore the reaction to make up for it to some degree.
HOAt, I am yet to review but molbase has its synthesis routes. HOBT will almost certainly work.
Still don't know how to find out how much to use though, don't know how stoichiometry works for peptides. - Often people use an excess of the cheapest
reagent to drive the reaction to completion, on resins, you use an excess of the reagents to drive the reaction to completion, but in many cases only
a small excess is needed, if careful, dry, and willing to wait a while longer. In solution, I aim at no more than 1.1 to 1.2 equivalents of the less
precious reagent, but sometimes add 2-3 eq of coupling reagent and base (Hunig's base or TEA commonly used.)
I hope ruthenium could be retrieved from used peptides and reactions because it is expensive. Only one step uses Grubbs, so not a big deal.
|
|
|
ziqquratu
Hazard to Others
  
Posts: 385
Registered: 15-11-2002
Member Is Offline
Mood: No Mood
|
|
If I recall correctly, the problem with Grubbs II is that it's remarkably unstable due to the nature of the intermediates in the catalytic cycle. From
memory, it's got an effective half life measured in hours, so you can't really extend the time too much. This is why it is typically used with such a
high loading (5-10 mol%), compared to many other commonly used catalysts.
There are alternative reagents - such as the Hoveyda-Grubbs catalysts - that are more stable; but also quite a bit more expensive.
In order to get the very best out of low loadings, you're best off to make sure that your reaction is as dry as possible, and to completely exclude
oxygen - for small scale, freeze-pump-thaw degas your solvents; for larger scale, sparge with dry nitrogen or argon for at least half an hour
(preferably longer) before adding the catalyst.
All that said, though, let me again emphasise - your compound of interest has a high MW, so even 5 mol% isn't going to be very much catalyst! For 1
mmol of peptide (586mg), at 5 mol%, you'd need 41 mg of Grubbs II (MW = 822).
|
|
|
SupaVillain
Hazard to Others
  
Posts: 171
Registered: 28-10-2014
Member Is Offline
Mood: No Mood
|
|
Hmmm i might buy hoveyda grubb2nd gen but not if it's only available from sigma since Im not a business. According to a paper it is only 50-100
micromoles per passage of cells, which is each time cells are removed so that new ones can grow and fill the petri dish back up. Some things really
are best prepared. Im trying to figure out how peptide stoichiometry works now.
Oh.
|
|
|
ziqquratu
Hazard to Others
  
Posts: 385
Registered: 15-11-2002
Member Is Offline
Mood: No Mood
|
|
SupaVillain, again, don't get hung up on "peptide stoichiometry". 1 mmol of amine plus 1 mmol of carboxylic acid plus 1 mmol coupling reagent(s) leads
to 1 mmol of amide. Everything else is Le Chatlier to force it to the right!
Our routine for peptide couplings on solid support was 1.5-2eq of Fmoc-amino acid (remember, the amine is attached to the resin and is the limiting
reagent), and 1.6-2.5eq of coupling reagent (a greater excess than of the amino acid). Typically in DMF, but DCM also works most of the time. If we
found that a particular coupling was failing to give good yields, we would usually do a "double coupling" - that is, do the coupling reaction twice
(so amine + 1.5 eq carboxylic acid; wash all that away and add another 1.5eq acid) - before moving on to deprotection and adding the following
residue. You could also use a larger excess, but double coupling is a more reliable way of boosting yields. After that, you can cap any unreacted
amine with acetic anhydride (for a short peptide, this might not be worthwhile, but it can simplify purification), then you remove the Fmoc group with
piperidine and move on to the next coupling.
|
|
|
|