IanCaio
Hazard to Self
 
Posts: 52
Registered: 26-9-2012
Member Is Offline
Mood: No Mood
|
|
Any Mopac7 users around?
Hello folks, been a while I don't post anything in the forums.
I've been following an intermediate organic chemistry course, which focus on the Molecular Orbital approach to chemical reactivity. Even though there
are still some doubts emerging from this course, which I plan to answer with time, I've been amazed with all the new concepts and how some of them
coincide with the image I had in my head about resonance/delocalization.
Of course I want to practice all those new concepts, so I looked for some free molecular orbital calculator to try visualizing some molecules orbitals
and predict reactivity or other properties.
I've ran into Mopac 7, which is opensource, compiled it on my notebook (which runs Ubuntu) and have been trying to figure how it works. So far it
seems to be a good way to calculate the LCAO coefficients, however I had some results that didn't seem to be right. Would be nice to have some more
experienced user to share information with and enlighten some doubts! 
So, anyone out there has some experience using Mopac 7?
Link to the Mopac 7 source (which I've used):
https://sourceforge.net/projects/mopac7/
|
|
|
Marvin
National Hazard
   
Posts: 995
Registered: 13-10-2002
Member Is Offline
Mood: No Mood
|
|
I've not used mopac for years. In fact when I last used it properly I had to type connection angles and distances into a BBC model B which submitted
jobs to a VAX. Throw us some examples of the successes and failures and maybe we can see the issue.
|
|
|
Polverone
Now celebrating 21 years of madness
        
Posts: 3186
Registered: 19-5-2002
Location: The Sunny Pacific Northwest
Member Is Offline
Mood: Waiting for spring
|
|
I have some stalled work to integrate Mopac7 in a framework with other semiempirical and ab initio quantum chemistry software here: https://github.com/mattbernst/polyhartree/blob/master/adapte... (see full project too)
Mopac7 is pretty buggy. If you want to run semiempirical calculations for important uses I suggest getting a MOPAC2016 license from http://openmopac.net/ (free for not-for-profit academic use). Or you could try using the pDynamo code that is also integrated in Polyhartree, but
that's a fair bit more work to set up.
For background and ancient source code for semiempirical calculations, see <a
href="http://library.sciencemadness.org/library/books/approximate_mo_theory.pdf">Approximate Molecular Orbital Theory</a> by Pople and
Beveridge.
PGP Key and corresponding e-mail address
|
|
|
IanCaio
Hazard to Self
 
Posts: 52
Registered: 26-9-2012
Member Is Offline
Mood: No Mood
|
|
Hello Marvin!
When I compiled the Mopac, there was a test file which I ran. It gave a theoretical heat of formation and the output of the program was supposed to be
the same. It was, so I'm assuming the program is running the way it's supposed to. However I had some weird results when trying to calculate some
molecular orbitals.
I'll make a step by step of a H2O MO calculation:
1 - Create a '.dat' file with the molecules and positions. I did this on Avogadro, exported as a '.mop' file and changed the file extension and some
keywords on a text editor. Here I found out that the initial geometry you set can result in some errors. I had the "CALCULATION ABANDONED AT THIS
POINT" error in a few tests, which was related to the geometry optimization. As far as I understood it means that in one of the iterations 3 atoms
aligned and this screws up with some coordinate system of Mopac. But I could fix that by defining some different starting position for the atoms and
trying again.
The 'water.dat' file:
| Code: | LARGE CHARGE=0 SINGLET PRECISE VECTORS
Water
O 0.000000 1 0.000000 1 0.000000 1 0 0 0
H 0.818040 1 0.000000 1 0.000000 1 1 0 0
H 2.229961 1 137.144343 1 0.000000 1 1 2 0 |
First line defining the Keywords:
LARGE - Results in a more detailed output as far as I understood
CHARGE=0 - Sets the molecule charge to 0
SINGLET - Make calculations for the molecule in the singlet state (I'm still not familiarized with the singlet/triplet state concept, but I saw in
some example water being treated as singlet)
PRECISE - Get more precises results using some 'tighter' criteria for finishing calculations
VECTORS - Shows calculated Eigenvectors and Eigenvalues. From what I understood, only from the (HOMO-9) up to the (LUMO+7), which sounds reasonable,
since they are the most important orbitals for molecular reactivity. I can also get the other orbitals Eigenvectors and Eigenvalues through the DEBUG
keyword, which will print every update on all the orbitals Eigenvectors/values.
2 - I go to Linux terminal, reach the Mopac7 folder and run the following command:
| Code: | ./run_mopac7 MyMopacWorks/Water/water |
Which will run mopac7 parsing the water.dat file I created as an argument.
3 - The following output is printed:
| Code: |
starting mopac7 job MyMopacWorks/Water/water
AN UNOPTIMIZABLE GEOMETRIC PARAMETER HAS
BEEN MARKED FOR OPTIMIZATION. THIS IS A NON-FATAL ERROR
*******************************************************************************
** MOPAC FOR LINUX (PUBLIC DOMAIN VERSION) MTA ATOMKI, Debrecen, 95-JUN-21 **
*******************************************************************************
MNDO CALCULATION RESULTS
*******************************************************************************
* MOPAC: VERSION 7.01 CALC'D. Thu Jul 7 12:30:15 2016
* VECTORS - FINAL EIGENVECTORS TO BE PRINTED
* LARGE - EXPANDED OUTPUT TO BE PRINTED
* SINGLET - SPIN STATE DEFINED AS A SINGLET
*
*
*
* CHARGE ON SYSTEM = 0
*
*
*
* T= - A TIME OF 3600.0 SECONDS REQUESTED
* DUMP=N - RESTART FILE WRITTEN EVERY 3600.0 SECONDS
* PRECISE - CRITERIA TO BE INCREASED BY 100 TIMES
***********************************************************************060BY060
LARGE CHARGE=0 SINGLET PRECISE VECTORS
Water
ATOM CHEMICAL BOND LENGTH BOND ANGLE TWIST ANGLE
NUMBER SYMBOL (ANGSTROMS) (DEGREES) (DEGREES)
(I) NA:I NB:NA:I NC:NB:NA:I NA NB NC
1 O
2 H .81804 * 1
3 H 2.22996 * 137.14434 * 1 2
CARTESIAN COORDINATES
NO. ATOM X Y Z
1 O .0000 .0000 .0000
2 H .8180 .0000 .0000
3 H -1.6347 1.5167 .0000
MOLECULAR POINT GROUP : CS
H: (MNDO): M.J.S. DEWAR, W. THIEL, J. AM. CHEM. SOC., 99, 4899, (1977)
O: (MNDO): M.J.S. DEWAR, W. THIEL, J. AM. CHEM. SOC., 99, 4899, (1977)
RHF CALCULATION, NO. OF DOUBLY OCCUPIED LEVELS = 4
INTERATOMIC DISTANCES
0
O 1 H 2 H 3
------------------------------------------
O 1 .000000
H 2 .818040 .000000
H 3 2.229961 2.883824 .000000
CYCLE: 1 TIME: .00 TIME LEFT: 3600.0 GRAD.: 77.347 HEAT: 115.6750
CYCLE: 2 TIME: .00 TIME LEFT: 3600.0 GRAD.: 80.583 HEAT: 108.9804
CYCLE: 3 TIME: .00 TIME LEFT: 3600.0 GRAD.: 91.399 HEAT: 101.8649
CYCLE: 4 TIME: .00 TIME LEFT: 3600.0 GRAD.: 103.971 HEAT: 93.30336
CYCLE: 5 TIME: .00 TIME LEFT: 3600.0 GRAD.: 116.919 HEAT: 84.09738
CYCLE: 6 TIME: .00 TIME LEFT: 3600.0 GRAD.: 129.894 HEAT: 74.37389
CYCLE: 7 TIME: .00 TIME LEFT: 3600.0 GRAD.: 253.668 HEAT: 48.79623
CYCLE: 8 TIME: .00 TIME LEFT: 3600.0 GRAD.: 186.294 HEAT: 10.71791
CYCLE: 9 TIME: .00 TIME LEFT: 3600.0 GRAD.: 173.676 HEAT:-25.94593
CYCLE: 10 TIME: .00 TIME LEFT: 3600.0 GRAD.: 73.171 HEAT:-45.79115
CYCLE: 11 TIME: .00 TIME LEFT: 3600.0 GRAD.: 33.460 HEAT:-60.38013
CYCLE: 12 TIME: .00 TIME LEFT: 3600.0 GRAD.: .794 HEAT:-60.93768
HEAT OF FORMATION TEST SATISFIED
PETERS TEST SATISFIED
-------------------------------------------------------------------------------
LARGE CHARGE=0 SINGLET PRECISE VECTORS
Water
PETERS TEST WAS SATISFIED IN BFGS OPTIMIZATION
SCF FIELD WAS ACHIEVED
MNDO CALCULATION
VERSION 7.01
Thu Jul 7 12:30:15 2016
FINAL HEAT OF FORMATION = -60.93808 KCAL
TOTAL ENERGY = -351.42483 EV
ELECTRONIC ENERGY = -500.46547 EV
CORE-CORE REPULSION = 149.04064 EV
IONIZATION POTENTIAL = 12.19190
NO. OF FILLED LEVELS = 4
MOLECULAR WEIGHT = 18.015
SCF CALCULATIONS = 32
COMPUTATION TIME = .004 SECONDS
ATOM CHEMICAL BOND LENGTH BOND ANGLE TWIST ANGLE
NUMBER SYMBOL (ANGSTROMS) (DEGREES) (DEGREES)
(I) NA:I NB:NA:I NC:NB:NA:I NA NB NC
1 O
2 H .94310 * 1
3 H .94330 * 106.72098 * 1 2
INTERATOMIC DISTANCES
0
O 1 H 2 H 3
------------------------------------------
O 1 .000000
H 2 .943103 .000000
H 3 .943301 1.513662 .000000
MOLECULAR POINT GROUP : C2V
EIGENVECTORS
Root No. 1 2 3 4 5 6
1 A1 1 B2 2 A1 1 B1 3 A1 2 B2
-40.037 -19.107 -14.475 -12.192 5.445 6.722
S O 1 .8837 .0001 .3545 .0000 -.3056 -.0003
Px O 1 .0856 -.5963 -.4933 .0000 -.3242 -.5373
Py O 1 .1151 .4434 -.6633 .0000 -.4370 .3990
Pz O 1 .0000 .0000 .0000 -1.0000 .0000 .0000
S H 2 .3151 -.4732 -.3090 .0000 .5520 .5260
S H 3 .3150 .4732 -.3091 .0000 .5530 -.5249
NET ATOMIC CHARGES AND DIPOLE CONTRIBUTIONS
ATOM NO. TYPE CHARGE ATOM ELECTRON DENSITY
1 O -.3253 6.3253
2 H .1626 .8374
3 H .1627 .8373
DIPOLE X Y Z TOTAL
POINT-CHG. .525 .706 .000 .879
HYBRID .539 .725 .000 .904
SUM 1.064 1.431 .000 1.783
CARTESIAN COORDINATES
NO. ATOM X Y Z
1 O .0000 .0000 .0000
2 H .9431 .0000 .0000
3 H -.2714 .9034 .0000
ATOMIC ORBITAL ELECTRON POPULATIONS
1.81324 1.21239 1.29968 2.00000 .83735 .83734
TOTAL CPU TIME: .00 SECONDS
== MOPAC DONE ==
|
First he seems to make a serie of iteractions to find the correct geometry, using the Heat of Formation as a controlling parameter. Once he reaches
the lowest Heat of Formation he assumes the geometry is right. The final Heat of Formation was -60.94Kcal/mol which is not really far from the
literature one (I found about -68.36Kcal/mol on the internet).
Now to the Eigenvectors list:
It seems to unconsider the 1s orbital from oxygen, which is understandable since it's much lower in energy in comparison with the 2-orbitals and would
probably have a small role interacting with those. It would be interesting though, that I could set it to be taken into account for bigger precision
on lower energy orbitals.
The HOMO sounds right, considering the highest energy orbital is the lone pair which is situated in the 2Pz orbital from Oxygen.
But when I tried visualizing the orbitals below HOMO, they looked quite weird. To visualize it, I had to pass those informations to a '.aux' file,
which was a little tricky for a newbie as me. I basically took an example '.aux' file and tried to change the parameters to the ones that I
calculated.
The final '.aux' file:
| Code: |
START OF MOPAC FILE
####################################
# #
# Start of Input data #
# #
####################################
MOPAC_VERSION=MOPAC2007.7.150W
DATE="Mon Jun 4 12:15:37 2007"
METHOD=PM6
TITLE=" Water"
KEYWORDS=" SYMMETRY"
ATOM_EL[0003]=
O H H
ATOM_X:ANGSTROMS[0009]=
0.0000 0.0000 0.0000
0.9431 0.0000 0.0000
-0.2714 -0.9034 0.0000
AO_ATOMINDEX[0006]=
1 1 1 1 2 3
ATOM_SYMTYPE[0006]=
S PX PY PZ S S
AO_ZETA[0006]=
5.4218 2.2710 2.2710 2.2710 1.2686 1.2686
ATOM_PQN[0006]=
2 2 2 2 1 1
NUM_ELECTRONS=0008
EMPIRICAL_FORMULA="H2 O"
####################################
# #
# Final SCF results #
# #
####################################
HEAT_OF_FORMATION:KCAL/MOL=-60.9381
ENERGY_ELECTRONIC:EV=-351.4248
ENERGY_NUCLEAR:EV=149.0406
POINT_GROUP=C2v
DIPOLE:DEBYE=+0.206870D+01
AREA:SQUARE ANGSTROMS=+0.424512D+02
VOLUME:CUBIC ANGSTROMS=+0.251444D+02
ATOM_X_OPT:ANGSTROMS[0009]=
0.0000 0.0000 0.0000
0.9431 0.0000 0.0000
-0.2714 -0.9034 0.0000
ATOM_CHARGES[0003]=
-0.3253 +0.1626 +0.1626
OVERLAP_MATRIX[000021]=
# Lower half triangle only
1.0000 0.0000 1.0000 0.0000 0.0000 1.0000 0.0000 0.0000 0.0000 1.0000
0.1618 0.2330 0.3180 0.0000 1.0000 0.1618 0.2330 -0.3180 0.0000 0.2333
1.0000
EIGENVECTORS[000036]=
0.8837 0.0856 0.1151 0.0000 0.3151 0.3150 0.0001 -0.5963 0.4434 0.0000
-0.4732 0.4732 0.3545 -0.4933 -0.6633 0.0000 -0.3090 -0.3091 0.0000 0.0000
0.0000 -1.0000 0.0000 0.0000 -0.3056 -0.3242 -0.4370 0.0000 0.5520 0.5530
-0.0003 -0.5373 0.3990 0.0000 0.5260 -0.5249
TOTAL_DENSITY_MATRIX[000021]=
# Lower half triangle only
1.7601 -0.3333 1.5370 0.0000 0.0000 1.3216 0.0000 0.0000 0.0000 2.0000
0.3944 0.5480 0.6695 0.0000 0.6907 0.3944 0.5480 -0.6695 0.0000 0.0123
0.6907
M.O.SYMMETRY_LABELS[0006]=
1a1 1b2 2a1 1b1 3a1 2b2
EIGENVALUES[0006]=
-40.037 -19.107 -14.475 -12.192 5.445 6.722
MOLECULAR_ORBITAL_OCCUPANCIES[00006]=
1.81324 1.21239 1.29968 2.00000 0.83735 0.83734
|
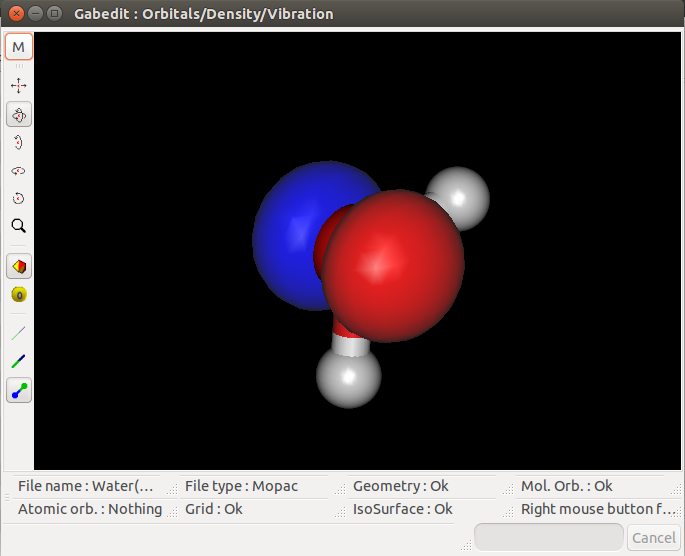
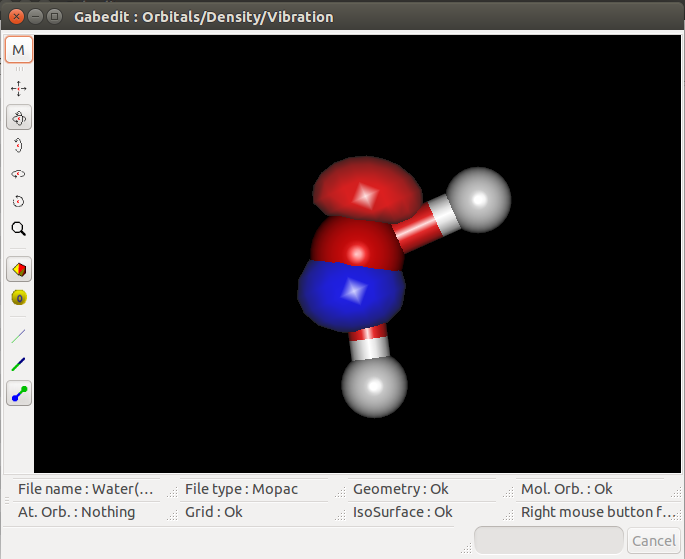
I then tried to visualize the MOs on Gabedit and had the following results for the HOMO, HOMO-1 and LUMO MOs:
HOMO:
HOMO-1:
LUMO:
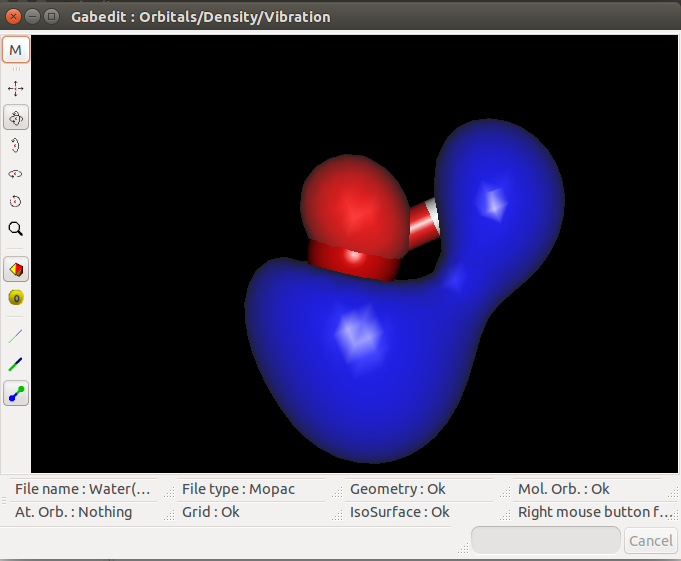
I don't know if the '.aux' file was properly edited, but the HOMO seems to fit the results from Mopac. However, the HOMO-1 seems a bit weird and the
LUMO even weirder (thought they would be symetric in relation to the hydrogens).
I'm sorry about the really long post, I just tried to be as detailed as possible about what I did and the results I had. Any ideas why some orbitals
doesn't seem to fit the theoretical ones?
Thanks in advance!
|
|
|
IanCaio
Hazard to Self
 
Posts: 52
Registered: 26-9-2012
Member Is Offline
Mood: No Mood
|
|
Hey Polverone!
I'll check it out, not sure if I'll be able to understand the work though  . Not
only I'm a little rusty in programming as I'm kind of new to quantum chemistry! . Not
only I'm a little rusty in programming as I'm kind of new to quantum chemistry!
I would Mopac mostly for educational purposes, trying to practice with the new concepts I'm learning right now. I'm not sure if they would give me an
MOPAC 2016 Academic License, but it's worth a try.
I just downloaded the book and will try getting a look at it! (Even though I think I lack the background to understand it yet!)
By the way, I've a MOPAC7 '.pdf' manual here, in case it interests anyone. Found it online and it has some good information about the functionalities
of Mopac7.
|
|
|
Polverone
Now celebrating 21 years of madness
        
Posts: 3186
Registered: 19-5-2002
Location: The Sunny Pacific Northwest
Member Is Offline
Mood: Waiting for spring
|
|
I believe that Avogadro's mopac generator is for the modern software available from openmopac.net. I wouldn't expect those files to work correctly
with Mopac7. When I started work on Polyhartree I couldn't find any free molecular editor that would generate Mopac7 files; I had to follow the Mopac7
manual and some trial and error experimentation to start generating correct inputs.
PGP Key and corresponding e-mail address
|
|
|
IanCaio
Hazard to Self
 
Posts: 52
Registered: 26-9-2012
Member Is Offline
Mood: No Mood
|
|
Indeed, the file generated from Avogadro is a '.mop' file, which is supposed to be used by the modern Mopac versions, but I believe the coordinates
system is the same. So I usually export the '.mop' file and edit it on a text editor, deleting the keywords that can't be understood on Mopac7 (like
AUX and PM6), and adding some keywords that are supposed to work on Mopac7 (like VECTORS, DEBUG, PRECISE and so on). I also change the file extension
from '.mop' to '.dat' which is the standard mopac7 input extension.
I only use Avogadro to get the coordinates from the atoms. The coordinate system is well explained on the Mopac7 manual, but I still didn't understand
it fully to do it by hand 
|
|
|