chemoleo
Biochemicus Energeticus
    
Posts: 3005
Registered: 23-7-2003
Location: England Germany
Member Is Offline
Mood: crystalline
|
|
Selective Coumarin modification or destruction
As the title says, I'd like to selectively destroy methoxy-coumarin, which is amide-bonded to a peptide.
Structure of coumarin (methoxylated in this case): (you can ignore the fluorenyl group, which is the triple ring system on lower left):

The idea is that coumarin, being a fluorophore, is selectively destroyed to either make it non-fluorescent, or to change its fluorescence properties,
i.e. excitation and emission spectra.
As the methoxy coumarin is on a peptide (i.e. containing any of the 20 natural amino acids it makes it more challenging, as there are numerous
reactive groups (-CH2OH (serine, threonine), RSH (cysteines), indole rings (tryptophan), phenolic rings (tyrosine), guanidyl groups (arginine), amines
(lysines) and carboxyl (glutamic, aspartic acid). Hmmm I can see myself this makes it difficult.
Anyway, if someone can suggest a way to selectively modify the coumarin, whilst not modifying any of the other groups present in a peptide, that would
be absolutely grand!
Never Stop to Begin, and Never Begin to Stop...
Tolerance is good. But not with the intolerant! (Wilhelm Busch)
|
|
|
The_Davster
A pnictogen
      
Posts: 2861
Registered: 18-11-2003
Member Is Offline
Mood: .
|
|
When you say we can ignore the fluorene system do you mean all associated reactivity there? If we cannot ignore its reactivity, all bases are out...
What about a mild reducing agent, I know one of the amino acids has a C=C double bond, but your coumarin's double bond is alpha to a carbonyl which
leaves it open to mild reducing conditions like sodium borohydride.
Should kill any emission and I don't think the peptides are in any danger this way, but I could be wrong. I know very little of amino acid chemistry
in general...
[Edited on 19-7-2008 by The_Davster]
|
|
|
chemoleo
Biochemicus Energeticus
    
Posts: 3005
Registered: 23-7-2003
Location: England Germany
Member Is Offline
Mood: crystalline
|
|
In the final peptide sequence, there is no Fmoc (fluorenyl) present, as Fmoc only serves as a base-labile protecting group during synthesis. So the
Lys-methoxy coumarin is amide coupled to the carboxy of the peptide chain. So yes, just pretend the Fmoc isn't even there - I just couldn't find a
structure of methoxy-coumarin on its own.
Here are the amino acids:

The coumarin double bond next to the carbonyl, isn't it resonance stabilised, giving rise to the red-shifted fluorescence?
But yes, if there's something that would split the cyclic ester, that would be perfect!
But there are other unsaturated rings systems in Histidine, Tyrosine and Tryptophan - please check them out - would they not be affected by NaBH4?
[Edited on 19-7-2008 by chemoleo]
Never Stop to Begin, and Never Begin to Stop...
Tolerance is good. But not with the intolerant! (Wilhelm Busch)
|
|
|
The_Davster
A pnictogen
      
Posts: 2861
Registered: 18-11-2003
Member Is Offline
Mood: .
|
|
Hmm, just noticed the C=N in histidine...I worry about this being reduced.
I imagine in the organic lit there has got to be something out there on the specific reduction of a C=C alpha to a carbonyl while leaving C=N, but I
don't know anything off the top of my head.
|
|
|
chemoleo
Biochemicus Energeticus
    
Posts: 3005
Registered: 23-7-2003
Location: England Germany
Member Is Offline
Mood: crystalline
|
|
Had an idea - I know that during Edman degradation the NH2-terminus of the peptides is attacked by isothiocyanate, and the very first amino acid of
the peptide is taken off.
Since the coumarin is at the *end* of the peptide, there is a free -COOH, which could be used for degradation of amino acids at the C-terminus!
Indeed there's a thing called Bergmann degradation - Check this:
http://en.wikipedia.org/wiki/Bergmann_degradation

| Quote: |
Bergmann degradation is series of chemical reactions designed to remove a single amino acid from the carboxylic acid end of a peptide.
The acyl azide of a peptide (1) undergoes a Curtius rearrangement in the presence of benzyl alcohol (2) to give a benzyl carbamate (3). The Cbz group
of intermediate 3 is removed by hydrogenolysis to give an unsubstituted amide (4) and an aldehyde (5). |
Question, how do I make the acylazide without acylating other things? I guess I need to dig up the 1934 Science paper.
1. ^ Bergmann, M. Science 1934, 79, 439.
2. ^ Bergmann, M.; Zervas, L. J. Biol. Chem. 1936, 113, 341.
http://www.jbc.org/cgi/reprint/113/2/341
Edit:
Never mind this particular degradation, it won't work here - the scales are on the order of a few mg, so multiple chemical transformations (as would
be required by the Bergmann degradation) won't be possible.
Something simple and effective would be best.
[Edited on 19-7-2008 by chemoleo]
Never Stop to Begin, and Never Begin to Stop...
Tolerance is good. But not with the intolerant! (Wilhelm Busch)
|
|
|
matei
Hazard to Others
  
Posts: 205
Registered: 16-9-2006
Member Is Offline
Mood: No Mood
|
|
chemoleo,
On Gigapedia you can find a book called "Fluorescent and Luminescent Probes for Biological Activity, 2nd Ed." and another one "Dynamic Studies in
Biology - Phototriggers, Photoswitches and Caged Biomolecules". Maybe you'll find there something that inspires you!
|
|
|
chemoleo
Biochemicus Energeticus
    
Posts: 3005
Registered: 23-7-2003
Location: England Germany
Member Is Offline
Mood: crystalline
|
|
Some coumarin-labelled peptide was heated to 80 deg C in 5 M HCl for a few hours - not only did the fluorescene go down, but the solution turned a
violet/blue!!!!
It is due to the heat, an unheated solution remains stable.
Can anyone suggest what's happening? Ring opening of the lactone, sure, but the blue colour? Seems unlikely that the resultant vinyl acid would have
such a strong absorption, even in the presence of the phenyl ring!
Ideas anyone?
How could I enhance/speed up that process?
[Edited on 20-7-2008 by chemoleo]
Never Stop to Begin, and Never Begin to Stop...
Tolerance is good. But not with the intolerant! (Wilhelm Busch)
|
|
|
Nicodem
Super Moderator
      
Posts: 4230
Registered: 28-12-2004
Member Is Offline
Mood: No Mood
|
|
I would guess heating a peptide in 5M HCl at 80°C breaks down a few amide links or is your peptide so robust as to survive that? The violet/blue
color could be due to anything since at such conditions lots of condensation products can form which can be intensively colored even in trace amounts.
Coumarins are sensitive toward nucleophiles and one way you could try to selectively block its fluorescence could be by hydrazinolysis. Unfortunately
hydrazine could not be particularly well tolerated by your peptide, but it surely is worth trying. For the beginning, try adding your peptide in 1M
hydrazine hydrate solution in an appropriate solvent (water, methanol, NMP, dimethylacetamide or wherever it can dissolve it - just keep in mind that
a lot of solvents react with hydrazine). Monitor if the fluorescence changes in a couple of hours. If it does not, then try with heating.
If your peptide is not compatible with hydrazine, you can try with milder N-nucleophiles like piperidine or dimethylamine, but here you will
almost certainly have to heat above 50°C for any reaction.
…there is a human touch of the cultist “believer” in every theorist that he must struggle against as being
unworthy of the scientist. Some of the greatest men of science have publicly repudiated a theory which earlier they hotly defended. In this lies their
scientific temper, not in the scientific defense of the theory. - Weston La Barre (Ghost Dance, 1972)
Read the The ScienceMadness Guidelines!
|
|
|
chemoleo
Biochemicus Energeticus
    
Posts: 3005
Registered: 23-7-2003
Location: England Germany
Member Is Offline
Mood: crystalline
|
|
Now I think there's more than 1 reaction going on.
The colour changed from blue to green with time (while heated)
Also, in H2SO4 the colour is red, and stays that way.
Also, the emission redshifts by 80 nm at least- quite impressive!
Quite sad that the mass spec of the peptide was quite predictably bad, no original peptide was left  Am impressed that acid-catalysed amide bond cleavage works that well. Am impressed that acid-catalysed amide bond cleavage works that well.
One thing left to test is the initial blue product found with HCl, before peptide degradation sets in.
Thanks for the advice, Nicodem. I heard about hydrazinolysis, but more in the context of breaking amide bonds- is the ester bond in the lactone ring
more sensitive than amides? I'll try this nonetheless.
Here http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=11... it suggests to include 1% hydrazine sulfate, too, and a temp. of 100 deg C. But
will do a proper literature search tomorrow.
Can you possibly suggest a source for hyrazinolysis, or on the other nucleophiles you suggested?
Edit: On the matter of piperidine - don't think this would work, as 20% piperidine in DMF is used as a deprotecting reagent - straight on the nascent
peptide chain - and the coumarin is certainly stable under these conditions, even when the deprotection reaction is done at 80 deg C.
[Edited on 22-7-2008 by chemoleo]
Never Stop to Begin, and Never Begin to Stop...
Tolerance is good. But not with the intolerant! (Wilhelm Busch)
|
|
|
Nicodem
Super Moderator
      
Posts: 4230
Registered: 28-12-2004
Member Is Offline
Mood: No Mood
|
|
Interesting that even at 80°C there is no reaction between piperidine and your coumarin labeled peptide. Though I would tend to think piperidine,
pyrrolidine and dimethylamine would require pretty high temperatures for complete reaction with coumarins (like 100°C and more), I would nevertheless
expect at least some of the corresponding cynnamoyl amide to form already at 80°C. I never worked with peptides, but somehow I imagine you need LC-MS
to follow the reactions. In such case you would surely have noticed some such product at the corresponding m/z if it would have formed. I hope this is
no indication that the coumarin is protected inside a steric pocket of the peptide, as this could mean a slower reaction with hydrazine as well
(though hydrazine is much smaller nucleophile).
A glance at SciFinder gives a few references of the reaction of coumarins with hydrazine. For more results I would rather suggest a search in
Beilstein as that should give more hits. One example:
| Quote: | Reactions with coumarin: synthesis and reactions of coumarin sulfonamides.
Abdel-Bary, Hamed M.
Afinidad, 55 (1998) 67-71.
Abstract
Coumarin-6-sulfonyl chloride was aminated with different secondary amines to give the sulfonamides. Treatment of these with hydrazine under
controlled conditions effected ring-opening of the lactone ring to afford the corresponding o-hydroxycinnamoyl hydrazides which were converted to
hydrazones by reation with various aldehydes. The hydrazones were cyclized using acetic anhydride to yield oxadiazolines. Reaction of the hydrazides
with 4-toluoyl chloride afforded the corresponding N-toluoyl derivs. which cyclized with POCl3 to the corresponding 1,3,4-oxadiazole derivs.
Thiosemicarbazide derivs. were obtained by treatment of the hydrazides with PhNCS. Cyclization of the thiosemicarbazides using POCl3 afforded the
corresponding 1,3,4-thiadiazoles. |
The hydrazinolysis of amides is nearly not so rapid as the same reaction with esters, so in practice it should be possible to selectively destroy the
coumarin fragment without affecting the peptide part of the molecule. It is a matter of finding the appropriate conditions. So, it is best to start
with mild conditions and finding a method to monitor the reaction progress (like monitoring the change in fluorescence peak frequency or with LC-MS if
you have access to it).
The reactivity of the coumarin carbonyl is also higher than that of normal esters (due to coumarins being aryl esters and having an
alpha,beta-unsaturation they are considerably more electrophilic regardless of the stabilization due to the cyclic structure and conjugation). This is
also demonstrated by the following reaction where the normal ester group (-CH2COOBu) does not end up hydrazinolyzed while the coumarin opens up
forming the corresponding cinnamoyl hydrazide (from Acta Chimica Hungarica, 128 (1991) 35-40 and Indian Journal of Chemistry, Section
B 29B, (1990) 239-243):
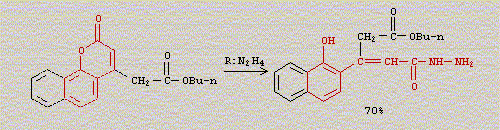
…there is a human touch of the cultist “believer” in every theorist that he must struggle against as being
unworthy of the scientist. Some of the greatest men of science have publicly repudiated a theory which earlier they hotly defended. In this lies their
scientific temper, not in the scientific defense of the theory. - Weston La Barre (Ghost Dance, 1972)
Read the The ScienceMadness Guidelines!
|
|
|
chemoleo
Biochemicus Energeticus
    
Posts: 3005
Registered: 23-7-2003
Location: England Germany
Member Is Offline
Mood: crystalline
|
|
Success!
I'll be darned!
After all the treatments the peptides received, hydrazine seems to have been successful!
The peptide, at 0.2 micromolar, was mixed with hydrazine hydrate, at 0.1 millimolar, in 50 mM NH4HCO3.
Aqueous conditions were chosen because hydrazine should be a strong enough nucleophile regardless the competition by the weaker OH nucleophile or even
reaction products (water).
Fluorescence was monitored with time, at room temp, but under constant irradiation of UV (325 nm) - so it is possible that the UV light catalysed the
reaction, that is why a similar reaction was run in the absence of UV (to be analysed later).
Anyway,
at t = 0 the mass was 2106
at t = 1900 sec not only did a small peak appear at 2139, but one almost at twice the mass, 4210 (+/-3 or so)
at t = 23500, the 2106 peak was tiny, whilst the 2139 peak was dominating the mass spectrum in this range, with an equally sized peak at 4210 (and a
smaller peak at 4125, and a few other tids and bits which I don't think are significant).
Now, 2139-2106 = 33
The hydrazine reacts with the ester to form a hydrazone:
R-O-CO-CH=R ---> ROH + NH2-NH-CO=R
Mass gained: 14+14+3+1 = 32 --> that's close enough for me (33 expected mass)!
Also, all fluorescence is lost! Remarkable; none of the acids/bases tried so far had such a drastical effect (they just caused a spectral shift of
fluorescence, but not loss of it)!
Now to the dimeric species:
4212 would be the mass if it was a conjugated dimer, without mass loss, but the mass is quite persistently at 4210.
The only dimerisation that could makes sense, within the right mass perimeter, would be R-O-C(-CH=R)(=N-N=)C(-CH=R)-O-R
Don't think that's right though, I've never heard of hydrazine attacking the C=O of an ester (that only happens with free carbonyls to my knowledge).
Also, the mass if off by 2 Dalton (which is admittedly well within the error of the mass spectrometer).
Any ideas what this dimeric species could be? How could I convert it to the monomeric one?
Also, I'd like to chemically make the hydrazone species of the 2139 mass less reactive, for instance, turn it into an amide. How?
I'm starting to enjoy this!
Never Stop to Begin, and Never Begin to Stop...
Tolerance is good. But not with the intolerant! (Wilhelm Busch)
|
|
|
Nicodem
Super Moderator
      
Posts: 4230
Registered: 28-12-2004
Member Is Offline
Mood: No Mood
|
|
I'm glad it worked.
Does your peptide have any mercapto groups (cysteine fragment)? If so then the dimer could be the consequence of peptide oxidation perhaps from air
oxygen in combination to UV irradiation. Such dimeric peptide would have the m/z of 4210. Is there any small peak at 4242 (2*M+N2H4-H2) and 4274
(2*M+2*N2H4-H2)? I suppose you did not used a buffer with dithiothreitol or whatever is it that you biochemists use to prevent disulfide formation?
Currently I have no other idea what that dimer could be. If your peptide does indeed contain cysteine, then treat it with dithiothreitol and do the MS
again and we'll see if this is it.
| Quote: | Originally posted by chemoleo
Also, I'd like to chemically make the hydrazone species of the 2139 mass less reactive, for instance, turn it into an amide. How?
I'm starting to enjoy this! |
It's never good enough... 
Hydrazides are not particularly reactive. They are still somewhat nucleophilic and can react with some oxidants, but the reactivity is more or less in
the range of an amino group which is ubiquitous in peptides anyway.
…there is a human touch of the cultist “believer” in every theorist that he must struggle against as being
unworthy of the scientist. Some of the greatest men of science have publicly repudiated a theory which earlier they hotly defended. In this lies their
scientific temper, not in the scientific defense of the theory. - Weston La Barre (Ghost Dance, 1972)
Read the The ScienceMadness Guidelines!
|
|
|
chemoleo
Biochemicus Energeticus
    
Posts: 3005
Registered: 23-7-2003
Location: England Germany
Member Is Offline
Mood: crystalline
|
|
No cysteines in there...just methionine.
The peptide otherwise contains serines, threonines, aspartate glutamate, glutamine, proline and asparagine, isoleucine and valine.
None of these are particularly reactive (compare with figure above)....so I'm pretty sure the dimerisation reaction must occur on the coumarin...
No peaks found at 4242 and 4274...
Could a preferential reaction occur with the free amides of asparagine or glutamine (as opposed to amide bond cleavage)? so that 2 R-CO-NH2 + H2NNH2
--> R-CO-NH-NH-CO-R + 2 NH3 ? Mass = 4210 - but then what's happening with the coumarin? The fluorescence is gone, and the main dimeric species
is at 4210.
Also I should potentially see even higher masses, trimeric, tetrameric etc species. Will check tomorrow (don't think I've seen higher masses).
Unless an glutamine/asparagine-CO-NH-NH2 preferentially attacks the lactone ring, then giving the 4125 peak (small but observable).
Any ideas?
Never Stop to Begin, and Never Begin to Stop...
Tolerance is good. But not with the intolerant! (Wilhelm Busch)
|
|
|
Drunkguy
Hazard to Others
  
Posts: 172
Registered: 23-12-2005
Member Is Offline
Mood: somewhat pissed.
|
|
Any idea how to get the hydrazine hydrate in the first place?
|
|
|
UnintentionalChaos
International Hazard
    
Posts: 1454
Registered: 9-12-2006
Location: Mars
Member Is Offline
Mood: Nucleophilic
|
|
1) this thread is over a year old since the last response and your question has almost nothing to do with the original topic.
2)We have a massive thread on hydrazine with functional syntheses. You've been here since 2005! You should know better 
[Edited on 6-27-09 by UnintentionalChaos]
Department of Redundancy Department - Now with paperwork!
'In organic synthesis, we call decomposition products "crap", however this is not a IUPAC approved nomenclature.' -Nicodem
|
|
|
Sandmeyer
National Hazard
   
Posts: 784
Registered: 9-1-2005
Location: Internet
Member Is Offline
Mood: abbastanza bene
|
|
You can hydrogenate the double bond in the heterocycle with pd/c in quant. yield, see tetrahedron, 63(48), 12026-12036; 2007. you can then regenerate
the coumarin (also in quant. yield) from the lactone with some of the usual aromatization reagents like ddq or palladium. Your Fmoc should be
allright.
|
|
|